NOTE: We are working on migrating this site away from MediaWiki, so editing pages will be disabled for now.
Difference between revisions of "JBrowse Tutorial 2012"
m (adding AMI info) |
m (Text replace - "<javascript>" to "<syntaxhighlight lang="javascript">") |
||
| Line 124: | Line 124: | ||
In this case, we have specified all of our track configurations in <code>pythium-1.conf</code>. | In this case, we have specified all of our track configurations in <code>pythium-1.conf</code>. | ||
| − | <javascript>... | + | <syntaxhighlight lang="javascript">... |
"TRACK DEFAULTS": { | "TRACK DEFAULTS": { | ||
| Line 214: | Line 214: | ||
JBrowse can display quantitative data directly from a BigWig file on your web server. Simply place the BigWig file in a directory accessible to your web server, and add a snippet of configuration to JBrowse to add the track, similar to: | JBrowse can display quantitative data directly from a BigWig file on your web server. Simply place the BigWig file in a directory accessible to your web server, and add a snippet of configuration to JBrowse to add the track, similar to: | ||
| − | <javascript> | + | <syntaxhighlight lang="javascript"> |
{ | { | ||
"label" : "bam_coverage", | "label" : "bam_coverage", | ||
| Line 257: | Line 257: | ||
Then a simple faceted track selection configuration might look like: | Then a simple faceted track selection configuration might look like: | ||
| − | <javascript> | + | <syntaxhighlight lang="javascript"> |
trackSelector: { | trackSelector: { | ||
type: 'Faceted', | type: 'Faceted', | ||
Revision as of 20:17, 8 October 2012
This JBrowse tutorial was presented by Robert Buels at the 2012 GMOD Summer School.
To follow along with the tutorial, you will need to use AMI ID: ami-b7fa4dde, name: GMOD 2012 day 2 start, available in the US East (N. Virginia) region. See the GMOD Cloud Tutorial for information on how to get this AMI.
Contents
Prerequisites
These have already been set up on the VM image.
Optional, for generating images from Wiggle files:
- libpng12-0
- libpng12-dev
- a C++ compiler
Optional, for BAM files (setup.sh tries to install these for you in the JBrowse directory):
- samtools, and its dependency libncurses5-dev
- perl module: Bio::DB::SAM
Other prerequisites are installed by JBrowse automatically.
This is how they were installed: (don't do this)
$ sudo apt-get install libpng12-0 libpng12-dev build-essential libncurses5-dev
Make sure you can copy/paste from the wiki.
It's also very useful to know how to tab-complete in the shell.
JBrowse Introduction
How and why JBrowse is different from most other web-based genome browsers, including GBrowse.
More detail: paper
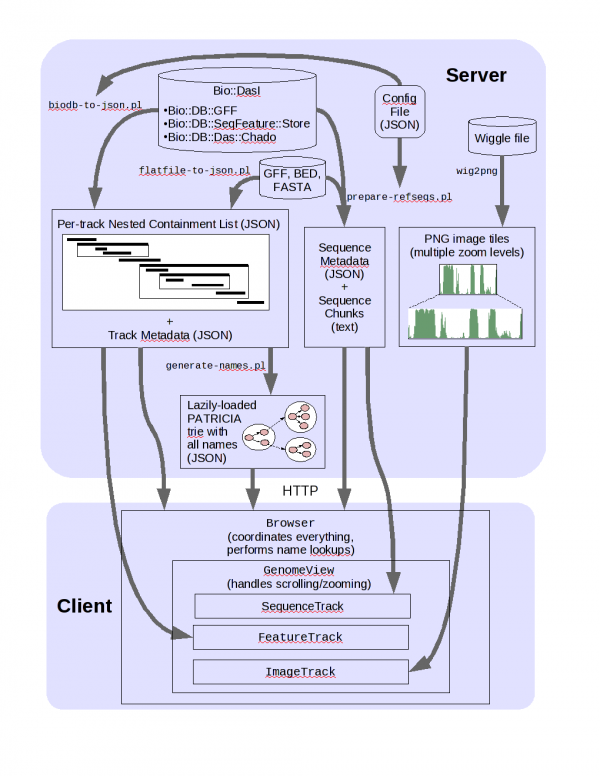
JBrowse Architecture
Setting up JBrowse
Getting JBrowse
- prepare a directory for JBrowse
$ cd /var/www $ sudo mkdir jbrowse_demo $ sudo chown ubuntu.ubuntu jbrowse_demo $ cd jbrowse_demo
- download the demo bundle from jbrowse.org and unzip it
$ wget http://jbrowse.org/info/GMOD_Aug_2012/GMOD_Summer_School_2012_JBrowse.zip $ unzip GMOD_Summer_School_2012_JBrowse.zip $ unzip JBrowse-1.6.0-min.zip $ mv JBrowse-1.6.0-min jbrowse
- run
setup.shto configure this copy of JBrowse
$ cd jbrowse $ ./setup.sh
Starting Point
Visit in web browser: http://ec2-##-##-##-##.compute-1.amazonaws.com/jbrowse_demo/jbrowse/
You should see just a blank white page.
Basic Steps
Setting up a JBrowse instance with feature data goes in three basic steps:
- Specify reference sequences
- Load feature data
- Index feature names
Data from a directory of files
Here, we'll use the Bio::DB::SeqFeature::Store adaptor in "memory" mode to read a directory of files. There are adaptors available for use with many other databases, such as Chado and Bio::DB::GFF.
Config file: pythium-1.conf
{
"description": "GMOD Summer School 2012 P. ultima Example",
"db_adaptor": "Bio::DB::SeqFeature::Store",
"db_args" : {
"-adaptor" : "memory",
"-dir" : ".."
},
...
Specify reference sequences
The first script to run is bin/prepare-refseqs.pl; that script is the way you tell JBrowse about what your reference sequences are. Running bin/prepare-refseqs.pl also sets up the "DNA" track.
Run this from within the jbrowse directory (you could run it elsewhere, but you'd have to explicitly specify the location of the data directory on the command line).
$ cd /var/www/jbrowse_demo/jbrowse $ bin/prepare-refseqs.pl --gff ../scf1117875582023.gff
Refresh it in your web browser, you should new see the JBrowse UI and a sequence track, which will show you the DNA base pairs if you zoom in far enough.
Load Feature Data
Next, we'll use biodb-to-json.pl to get feature data out of the database and turn it into JSON data that the web browser can use.
In this case, we have specified all of our track configurations in pythium-1.conf.
... "TRACK DEFAULTS": { "class": "feature" }, "tracks": [ { "track": "Genes", "key": "Genes", "feature": ["mRNA"], "autocomplete": "all", "class": "transcript", "subfeature_classes" : { "CDS" : "transcript-CDS", "UTR" : "transcript-UTR" }, "arrowheadClass" : "arrowhead" }, ... ]</javascript> <tt>track</tt> specifies the track identifier (a unique name for the track, for the software to use). This should be just letters and numbers and - and _ characters; using other characters makes things less convenient. <tt>key</tt> specifies a human-friendly name for the track, which can use any characters you want. <tt>feature</tt> gives a list of feature types to include in the track. <tt>autocomplete</tt> including this setting makes the features in the track searchable. <tt>urltemplate</tt> specifies a URL pattern that you can use to link genomic features to specific web pages. <tt>class</tt> specifies the [[Glossary#CSS|CSS]] class that describes how the feature should look. For this particular track, I've specified the <tt>transcript</tt> feature class. Run the <tt>bin/biodb-to-json.pl</tt> script with this config file to format this track, and the others in the file: $ <span class="enter">bin/biodb-to-json.pl --conf ../pythium-1.conf</span> Refresh JBrowse in your web browser. You should now see a bunch of annotation tracks. ==== Index feature names ==== When you generate JSON for a track, if you specify <tt>"autocomplete"</tt> then a listing of all of the feature names from that track (along with feature locations) will also be generated and used to provide feature searching and autocompletion. The <tt>bin/generate-names.pl</tt> script collects those lists of names from all the tracks and combines them into one big tree that the client uses to search. $ <span class="enter">bin/generate-names.pl -v</span> Visit in web browser, try typing a feature name, such as '''maker-scf1117875582023-snap-gene-0.26-mRNA-1'''. Notice that JBrowse tries to auto-complete what you type. === Data from flat files === We're going to add a couple more tracks that come from <tt>repeats.gff</tt>, a different flat file. ==== Features ==== To get feature data from flat files into JBrowse, use <tt>flatfile-to-json.pl</tt>. * We'll add a RepeatMasker track: $ <span class="enter">bin/flatfile-to-json.pl --trackLabel repeatmasker \ --type match:repeatmasker --getType --getSubfeatures --key RepeatMasker \ --arrowheadClass arrowhead --className generic_parent \ --subfeatureClasses '{"match_part" : "feature"}' --gff ../repeats.gff</span> * And then a RepeatRunner track: $ <span class="enter">bin/flatfile-to-json.pl --trackLabel repeatrunner \ --type protein_match:repeatrunner --getType --getSubfeatures \ --key RepeatRunner --arrowheadClass arrowhead --className generic_parent \ --subfeatureClasses '{"match_part" : "feature"}' --gff ../repeats.gff</span> Visit in web browser; you should see the two new RepeatMasker and RepeatRunner tracks. ==== BAM data ==== Now let's add some simulated short-read alignments from a BAM file. To import data from a BAM source: $ <span class="enter"> bin/bam-to-json.pl \ --bam ../simulated-sorted.bam \ --tracklabel BAM_data --key "BAM Data" === Quantitative data === ==== BigWig ==== JBrowse can display quantitative data directly from a BigWig file on your web server. Simply place the BigWig file in a directory accessible to your web server, and add a snippet of configuration to JBrowse to add the track, similar to: <syntaxhighlight lang="javascript"> { "label" : "bam_coverage", "key" : "BAM coverage", "storeClass" : "BigWig", "urlTemplate" : "../../simulated-sorted.bam.coverage.bw", "type" : "Wiggle", "variance_band" : true } </javascript> This can be added by either editing the <tt>data/trackList.json</tt> file with a text editor, or by running something like this at the command line to inject the track configuration: $ <span class="enter">echo ' { "label" : "bam_coverage", "key" : "BAM coverage", "storeClass" : "BigWig", "urlTemplate" : "../../simulated-sorted.bam.coverage.bw", "type" : "Wiggle", "variance_band" : true } ' | bin/add-track-json.pl data/trackList.json</span> ==== Tiled Images ==== JBrowse also has a formatter that converts wiggle-format data to image tiles. JBrowse does this with a C++ program, <code>setup.sh</code> attempts to compile for you. There isn't any Pythium wiggle example data for this class, but the command to make image tiles from a wiggle file takes the form: $ <span class="enter">bin/wig-to-json.pl --wig /path/to/wiggle.wig \ --tracklabel "coverage_wig" --key "Wiggle Coverage" --min 0 --max 50</span> === Faceted Track Selection === JBrowse has a new, very powerful faceted track selector that can be used to search for tracks using metadata associated with them. The track metadata is kept in a CSV-format file, with any number of columns, and with a "label" column whose contents must correspond to the track labels in the JBrowse configuration. The demo bundle contains an example <tt>trackMetadata.csv</tt> file, which can be copied into the <tt>data</tt> directory for use with this configuration. $ <span class="enter">cp trackMetadata.csv jbrowse/data</span> Then a simple faceted track selection configuration might look like: <syntaxhighlight lang="javascript"> trackSelector: { type: 'Faceted', }, trackMetadata: { sources: [ { type: 'csv', url: 'data/trackMetadata.csv' } ] } </javascript> The <tt>jbrowse_conf.json</tt> file in the <tt>jbrowse</tt> directory already conveniently contains this stanza, commented out. Uncomment it, refresh your browser, and you should now see the faceted track selector activated. == Upgrading an Existing JBrowse == If the old JBrowse is 1.3.0 or later, simply move the data directory from the old JBrowse directory into the new JBrowse directory. == Common Problems == * JSON syntax errors == Future JBrowse Plans == See the [[Media:JBrowse_gmod_aug2012.pdf|accompanying slides (PDF)]] == Other links == * Config file ref: http://jbrowse.org/code/jbrowse-master/docs/config.html * DIV test: http://jbrowse.org/test/boatdiv/boat.html [[Category:Tutorials]] [[Category:JBrowse]] [[Category:2012 Summer School]]