NOTE: We are working on migrating this site away from MediaWiki, so editing pages will be disabled for now.
Difference between revisions of "WebGBrowse"
| Line 3: | Line 3: | ||
{{ImageLeft|cgb_logo.png|WebGBrowse at the Center for Genomics and Bioinformatics||http://cgb.indiana.edu/}} | {{ImageLeft|cgb_logo.png|WebGBrowse at the Center for Genomics and Bioinformatics||http://cgb.indiana.edu/}} | ||
[http://webgbrowse.cgb.indiana.edu/ WebGBrowse] is a web interface for setting up [[GBrowse]] instances. It helps both biologists and bioinformaticians by allowing them to upload genome annotation information in [[GFF3]] format, configure display of each genomic feature through simple mouse clicks on a web browser and visualize the configured genome with the integrated GBrowse software. Thus, for biologists, WebGBrowse relieves both the burden of installing GBrowse and the need to learn the configuration semantics in order to setup their data on GBrowse. For bioinformaticians who are new to [[GBrowse]], WebGBrowse offers a headstart into understanding [http://webgbrowse.cgb.indiana.edu/glyphdoc.html GBrowse glyphs] and learning how to configure them. WebGBrowse is a good match for any organization that has users who want to create their own GBrowse instances, but lack enough computing resources and expertise to do so. WebGBrowse was developed at [http://www.iub.edu/ Indiana University's] [http://cgb.indiana.edu/ Center for Genomics and Bioinformatics]. WebGBrowse can be [http://webgbrowse.cgb.indiana.edu/software.html downloaded] and installed locally, or you can use the [http://webgbrowse.cgb.indiana.edu/ WebGBrowse at CGB]. | [http://webgbrowse.cgb.indiana.edu/ WebGBrowse] is a web interface for setting up [[GBrowse]] instances. It helps both biologists and bioinformaticians by allowing them to upload genome annotation information in [[GFF3]] format, configure display of each genomic feature through simple mouse clicks on a web browser and visualize the configured genome with the integrated GBrowse software. Thus, for biologists, WebGBrowse relieves both the burden of installing GBrowse and the need to learn the configuration semantics in order to setup their data on GBrowse. For bioinformaticians who are new to [[GBrowse]], WebGBrowse offers a headstart into understanding [http://webgbrowse.cgb.indiana.edu/glyphdoc.html GBrowse glyphs] and learning how to configure them. WebGBrowse is a good match for any organization that has users who want to create their own GBrowse instances, but lack enough computing resources and expertise to do so. WebGBrowse was developed at [http://www.iub.edu/ Indiana University's] [http://cgb.indiana.edu/ Center for Genomics and Bioinformatics]. WebGBrowse can be [http://webgbrowse.cgb.indiana.edu/software.html downloaded] and installed locally, or you can use the [http://webgbrowse.cgb.indiana.edu/ WebGBrowse at CGB]. | ||
| + | |||
| + | |||
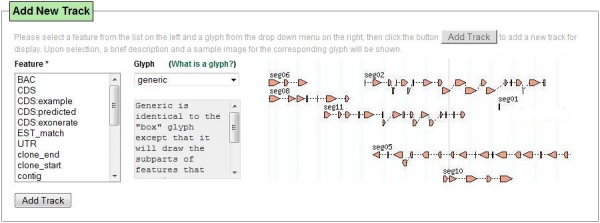
| + | [[Image:WebGBrowseAddTrack.jpg||600px|center|thumb|''Add new track'' dialog in WebGBrowse]] | ||
| + | |||
| + | |||
== Resources == | == Resources == | ||
| Line 10: | Line 15: | ||
* [http://webgbrowse.cgb.indiana.edu/glyphdoc.html GBrowse Glyph Library] - shows what all those [[GBrowse]] glyphs look like. | * [http://webgbrowse.cgb.indiana.edu/glyphdoc.html GBrowse Glyph Library] - shows what all those [[GBrowse]] glyphs look like. | ||
* [http://webgbrowse.cgb.indiana.edu/faq.html FAQ] - Frequently asked questions for WebGBrowse. | * [http://webgbrowse.cgb.indiana.edu/faq.html FAQ] - Frequently asked questions for WebGBrowse. | ||
| − | * [http://webgbrowse.cgb.indiana.edu/software.html | + | * Download source - Latest release from [http://webgbrowse.cgb.indiana.edu/software.html CGB] or latest development version from {{SF SVN|WebGBrowse/|SourceForge}}. |
* [http://bioinformatics.oxfordjournals.org/cgi/content/abstract/25/12/1550 "Podicheti R., Gollapudi R., Dong Q. (2009). WebGBrowse - a web server for GBrowse."] ''Bioinformatics'' 25(12): 1550-1551. - the WebGBrowse paper. | * [http://bioinformatics.oxfordjournals.org/cgi/content/abstract/25/12/1550 "Podicheti R., Gollapudi R., Dong Q. (2009). WebGBrowse - a web server for GBrowse."] ''Bioinformatics'' 25(12): 1550-1551. - the WebGBrowse paper. | ||
* [[January 2009 GMOD Meeting#WebGBrowse: GBrowse Configuration Management|"WebGBrowse: GBrowse Configuration Management"]], presentation by Ram Podicheti at the [[January 2009 GMOD Meeting]]. | * [[January 2009 GMOD Meeting#WebGBrowse: GBrowse Configuration Management|"WebGBrowse: GBrowse Configuration Management"]], presentation by Ram Podicheti at the [[January 2009 GMOD Meeting]]. | ||
| Line 22: | Line 27: | ||
{{MailingListsFor|WebGBrowse}} | {{MailingListsFor|WebGBrowse}} | ||
| − | |||
[[Category:GBrowse]] | [[Category:GBrowse]] | ||
| + | [[Category:GMOD Components]] | ||
Revision as of 19:44, 9 December 2010
- Mature release
- Active development
- Active support
WebGBrowse is a web interface for setting up GBrowse instances. It helps both biologists and bioinformaticians by allowing them to upload genome annotation information in GFF3 format, configure display of each genomic feature through simple mouse clicks on a web browser and visualize the configured genome with the integrated GBrowse software. Thus, for biologists, WebGBrowse relieves both the burden of installing GBrowse and the need to learn the configuration semantics in order to setup their data on GBrowse. For bioinformaticians who are new to GBrowse, WebGBrowse offers a headstart into understanding GBrowse glyphs and learning how to configure them. WebGBrowse is a good match for any organization that has users who want to create their own GBrowse instances, but lack enough computing resources and expertise to do so. WebGBrowse was developed at Indiana University's Center for Genomics and Bioinformatics. WebGBrowse can be downloaded and installed locally, or you can use the WebGBrowse at CGB.
Resources
Information on WebGBrowse is available in several places:
- User Tutorial - this is an extensive user guide for WebGBrowse
- GBrowse Glyph Library - shows what all those GBrowse glyphs look like.
- FAQ - Frequently asked questions for WebGBrowse.
- Download source - Latest release from CGB or latest development version from SourceForge.
- "Podicheti R., Gollapudi R., Dong Q. (2009). WebGBrowse - a web server for GBrowse." Bioinformatics 25(12): 1550-1551. - the WebGBrowse paper.
- "WebGBrowse: GBrowse Configuration Management", presentation by Ram Podicheti at the January 2009 GMOD Meeting.
WebGBrowse and GMOD
WebGBrowse was nominated for GMOD Membership on 2010/05/26. Feedback was universally favorable and we are now in the process of bringing WebGBrowse officially into GMOD.
Mailing Lists
| Mailing List Link | Description | Archive(s) | |
|---|---|---|---|
| WebGBrowse | gmod-webgbrowse | Questions, announcements, and development postings about the WebGBrowse front end to GBrowse configuration. | Nabble, Sourceforge |